
It's time to think BEyond dysplasia for Barrett's esophagus
The current paradigm for managing Barrett's esophagus is to wait and watch for dysplasia before considering treatment. That's a problem when dysplastic cells are difficult to find and tricky to diagnose.
What if there was a better way to identify BE patients with a high-risk of progression?
It's time to think BEyond dysplasia for Barrett's esophagus
The current paradigm for managing Barrett's esophagus (BE) is to wait and watch for dysplasia before considering treatment. That's a problem when dysplastic cells are difficult to find and tricky to diagnose.
What if there was a better way to identify BE patients with a high-risk of progression?
EAC is deadly, but you may be able help head it off at the pass
Esophageal cancer is deadly. BE is the only known clinical precursor to esophageal adenocarcinoma (EAC) and affects approximately 1 in 17 people in the United States. But a diagnosis of BE is important because BE is treatable if detected. In fact, 98% of dysplasia can be eradicated with radiofrequency ablation and EAC can effectively be prevented. The conundrum of BE is being able to identify which patients are most likely to progress to high-grade dysplasia or EAC.
The vast majority of patients are diagnosed with non-dysplastic Barrett's esophagus (NDBE), which is known to have a very low risk of progression (0.63% per year)1. But across a large population, thousands of these patients are going to progress.
That's why it shouldn't be surprising that 50% of BE patients in surveillance and who are later diagnosed with EAC had a prior diagnosis of NDBE— their population-based risk hid the reality that they had a much, much higher personal risk of progression to cancer. And worse, 25% of high-grade dysplasia and cancer diagnoses occur within 1 year of a prior endoscopy.
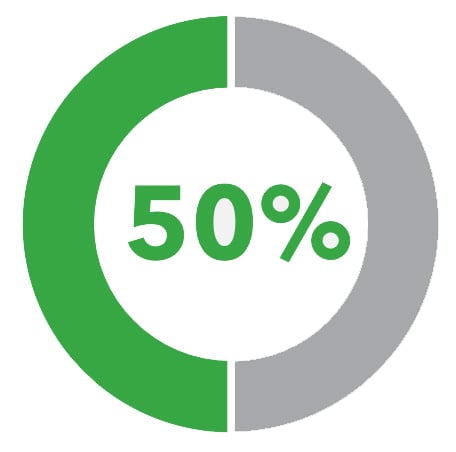
50% of annual HGD/EAC progressors in surveillance programs are initially diagnosed as non-dysplastic1
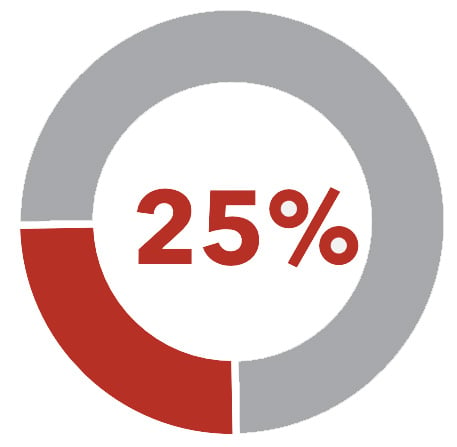
25% of high-grade/cancer diagnoses occur within one year of prior endoscopy2,3
To catch progressors earlier, BE patients need objective risk stratification that is adjunctive to histology.
Meet TissueCypher
TissueCypher is an AI-driven precision medicine test that helps predict the risk of progression to cancer in patients with Barrett’s esophagus. The test has the power to improve the standard of care for BE patients, many of whom are misclassified by traditional risk assessment.
The test has made it possible to assess molecular, cellular and morphological signatures in biopsies obtained during routine Barrett’s esophagus screening. The signatures identified by TissueCypher are powerful independent predictors of progression risk, and often precede the development of dysplasia, allowing for earlier identification, treatment, and management of truly high-risk BE patients.
Prognostic. Precise. Proven. Personalized.
TissueCypher is the first prognostic test using the elegance and precision of objective spatialomics to risk stratify patients with BE. The test has been proven in numerous published, peer-reviewed studies to help provide BE patients with a personalized probability of progression, to help allow for optimized treatment and care.The first EAC prognostic test, strongly evidenced with 14 peer-reviewed publications and studied in more than 8,000 patients

Independent analysis confirmed that TissueCypher is the strongest predictor of progression compared to clinical risk factors and pathology diagnosis
Actionable results, identify high-risk patients that progress at a rate equivalent to low-grade dysplasia and for whom increased surveillance or eradication therapy is appropriate
Learn more about the five foundational validation studies
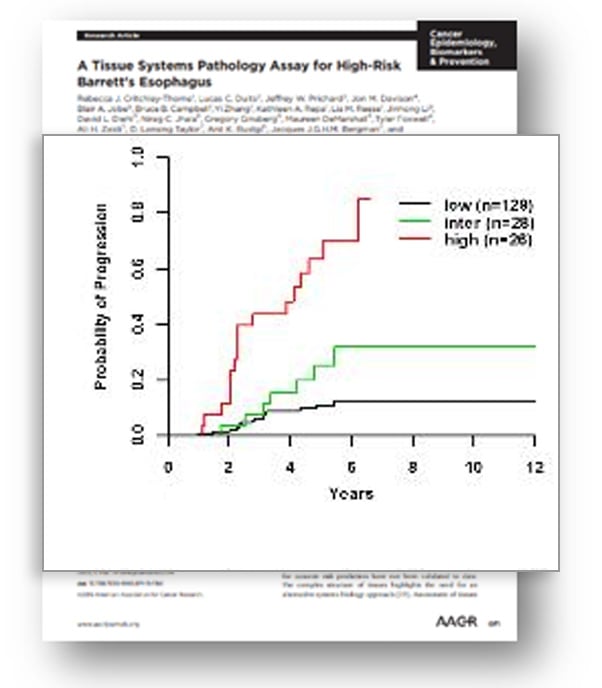
Incident progression from NDBE, IND, and LGD

Identifying missed prevalent progression
References
1 Progression rates: Rastogi et al. Gastrointest Endosc 2008; Krishnamoorthi et al. Gastrointest Endosc 2020; Singh et al. Gastrointest Endosc 2014; Wani et al. Clin Gastroenterol Hepatol 2011; Overholt et al. Gastrointest Endosc 2005.
2 Curvers et al. Am J Gastroenterol 2010.
3 Duits et al. Gut 2015.


